Multi-omics workflow for blood serum screening using GC×GC-TOFMS (Pierre-Hugues Stefanuto, MDCW 2024)
- Photo: MDCW: Multi-omics workflow for blood serum screening using GC×GC-TOFMS (Pierre-Hugues Stefanuto, MDCW 2024)
- Video: LabRulez: Pierre-Hugues Stefanuto: Multi-omics workflow for blood serum screening using GC×GC-TOFMS (MDCW 2024)
🎤 Presenter: Pierre-Hugues Stefanuto (Liege University, Liege, Belgium)
💡 Book in your calendar: 16th Multidimensional Chromatography Workshop (MDCW) 3 - 5. February 2025
Abstract
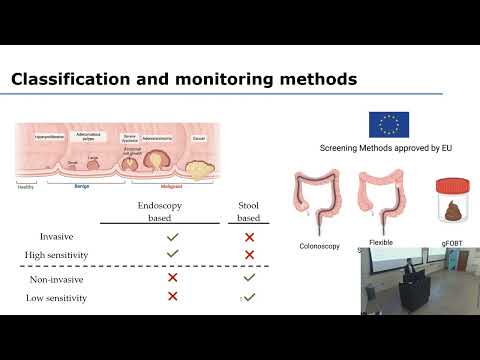
According to the world health organization (WHO), colorectal cancer (CRC) ranks as the third most frequently diagnosed cancer and the second leading cause of cancer-related deaths. The current endoscopic-based or stool-based diagnostic techniques are either highly invasive or lack sufficient sensitivity. Thus, there is a need for better screening approaches.
The rapid advancement of high throughput “omics” approaches, such as meta-genomics, transcriptomics, proteomics, metabolomics, lipidomics, microbiomics, and volatolomics, offer a potentially less invasive alternative than available techniques to develop novel biomarkers for colorectal cancer screening that could contribute to its clinical management.
For small molecules “omics” screening, gas chromatography coupled to mass spectrometry (GC-MS) represents a method of choice to screen biofluid samples. However, GC-MS is mostly used for targeted analysis due to the high complexity of the matrices. To tackle this limitation, comprehensive two-dimensional gas chromatography coupled to mass spectrometry provided increased separation capabilities, making it a method of choice for untargeted small molecule metabolomics.
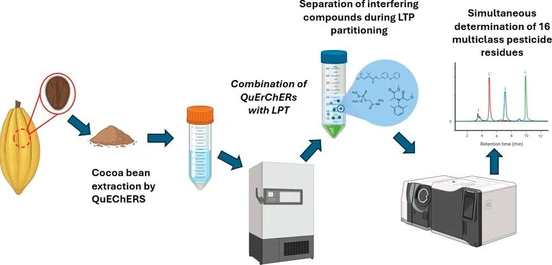
In this study, we analyzed 64 human serum samples representing three different groups of colorectal cancer using cutting edge GC×GC–LR/HR-TOFMS techniques. We analyzed samples with two different specifically tailored sample preparation approaches for lipidomics (fatty acids) (25 μL serum) and metabolomics (50 μL serum). In-depth chemometric screening with supervised and unsupervised approaches and metabolic pathway analysis were applied on both datasets. To ensure the high quality of the data produced, we have established a strict QAQC protocol including pooled sampled, serum SRM, and standard mixtures injections.
-Workshop-LOGO_s.webp)